Signal Strength Aware Latent Spaces Reveal Molecularly Distinct Substructures within Human Kidney Tissue
Published in Preprint, 2026
As datasets grow increasingly high-dimensional and complex, distinguishing a condensed set of interpretable underlying factors becomes essential. In spatial omics, for example, hundreds to thousands of molecular features per observation promise unprecedented biological insight. However, without meaningful latent representations, that potential remains markedly untapped.
We propose a new approach based on the beta-variational autoencoder and kernel density estimation to dissect data along independent, uncertainty-aware, and interpretable (yet non-linear) latent axes. We include a novel comparative-latent-traversal algorithm to translate latent findings back into the original measurement context. Demonstrating on imaging mass spectrometry-based molecular imaging of human kidney, the approach’s disentangling properties are shown to impress a latent space structure that separates signal strength from relative signal content, offering exceptional chemical insight. Our approach uncovers unexpected subdivisions within kidney proximal tubules, confirmed to be biological, and reveals hereto-unknown lipid species differentiating them. This confirms our workflow’s potential as an interpretation-and-hypothesis-generating discovery tool.
(a) Synthetic dataset generation \(\{x_i\}_{i\in I}\). 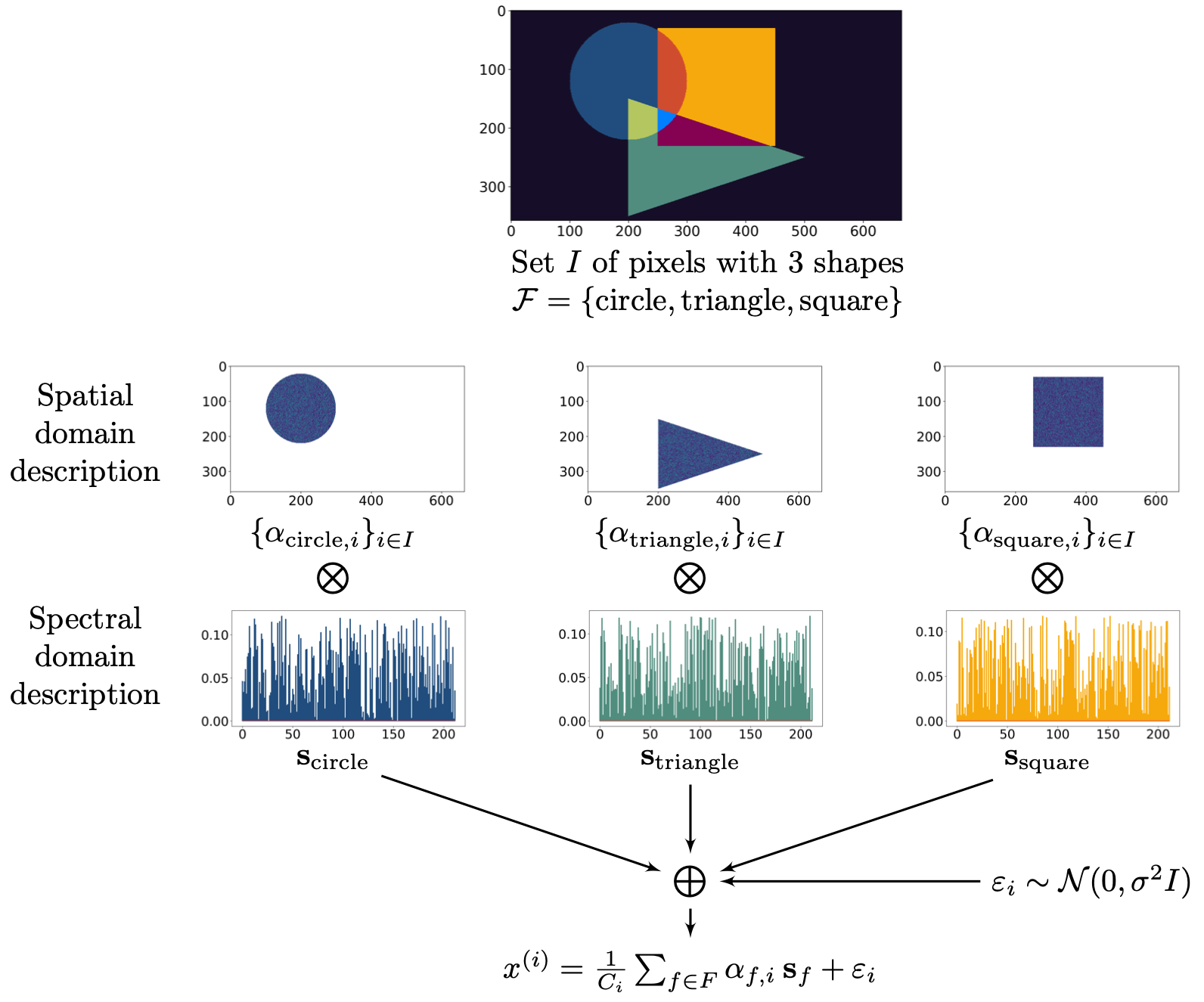 | |
(b) Latent representation colored by tissue structure membership. 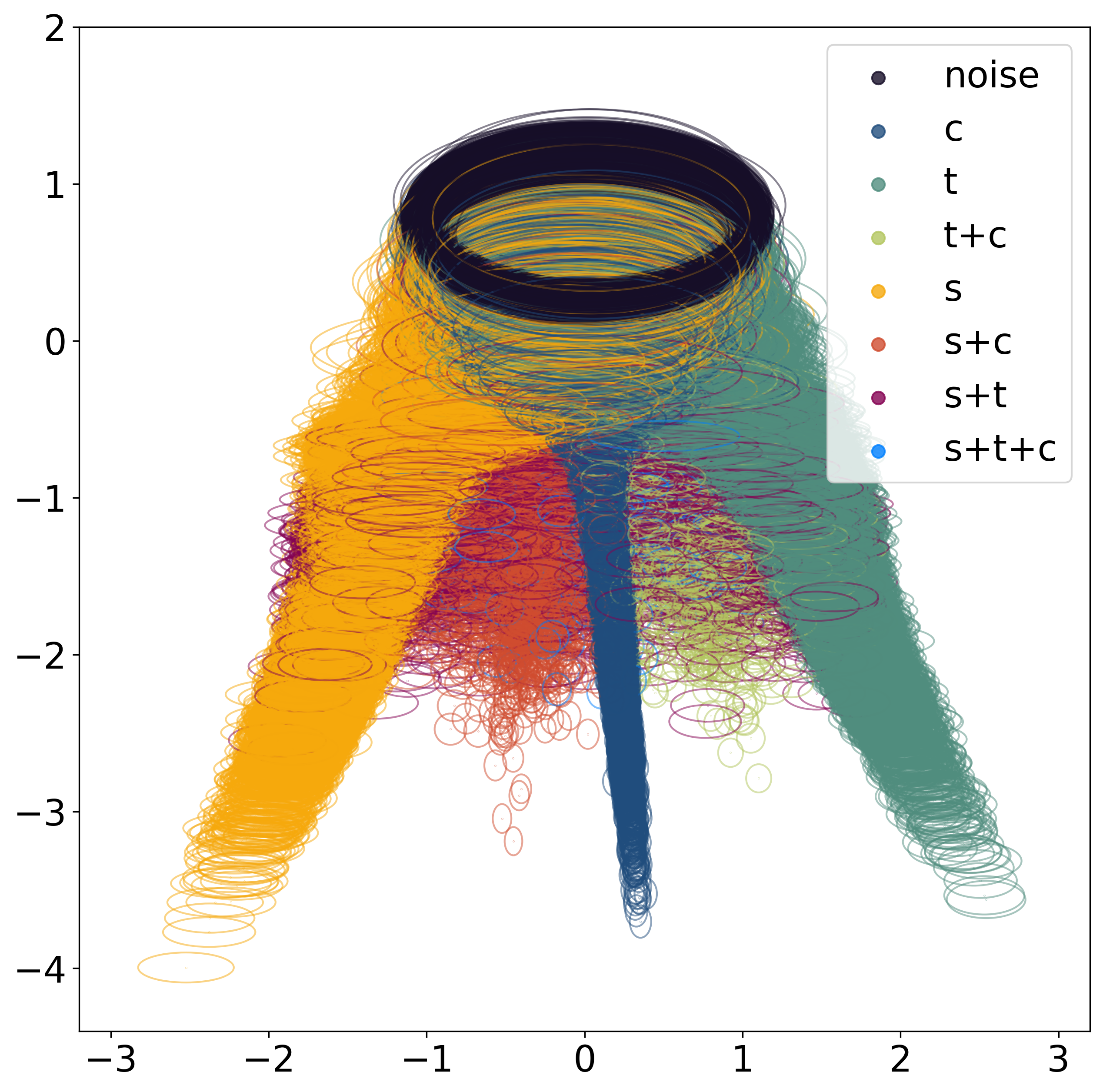 | (c) Latent representation colored by \(\text{SNR}_i = \frac{\|\sum_{f\in\mathcal{F}} \alpha_f \mathbf{s}_f\|^2}{\mathbb{E}[\|\epsilon_i\|^2]}\) 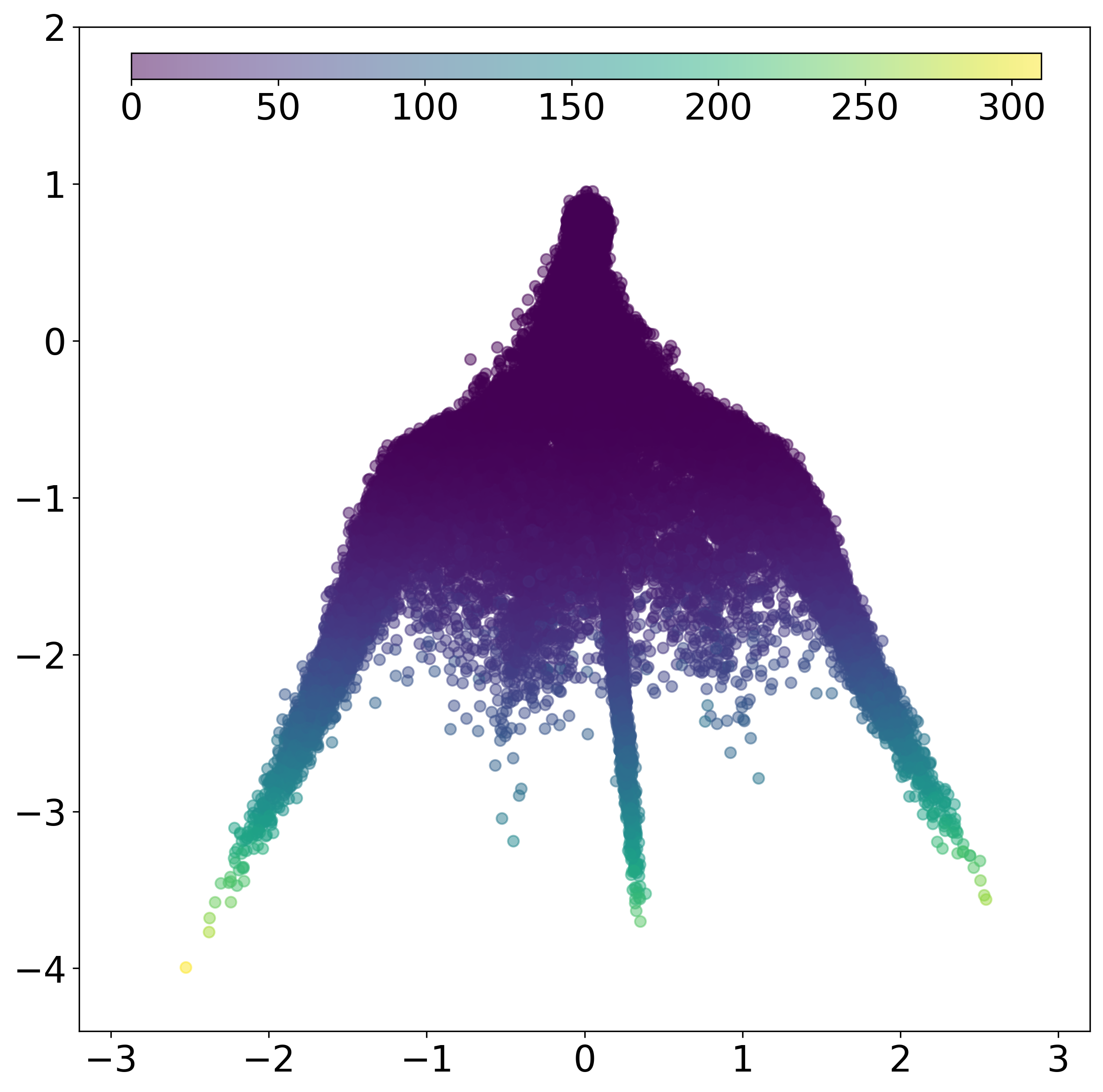 |
